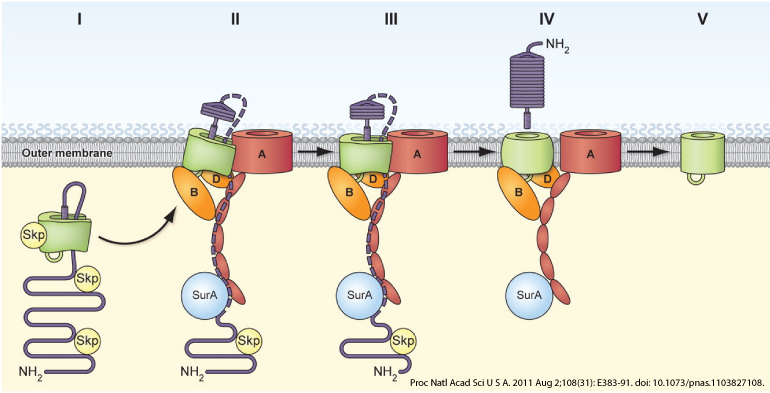
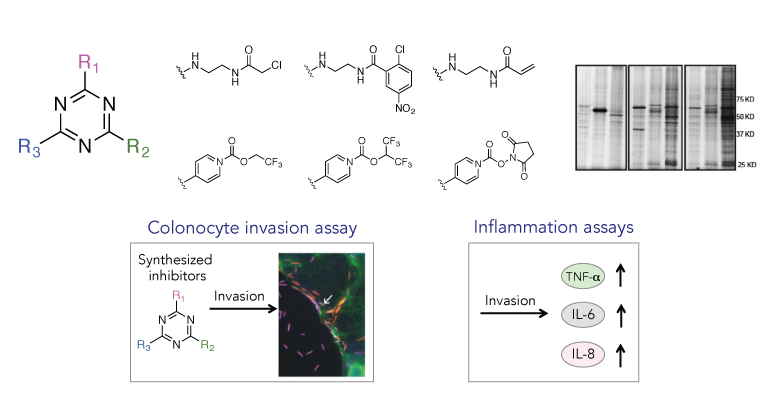
Autotransporters are type V secreted proteins that can be presented on the surface of bacteria, or secreted after being cleaved from the outer membrane. They are the largest family of bacteria virulence factors, and most have been characterized as adhesins and proteases. Previous studies have shown these proteins to be responsible for invoking an inflammatory response during infection. We will investigate if autotransporter adhesins and proteases could be triggering this inflammation in cancer.

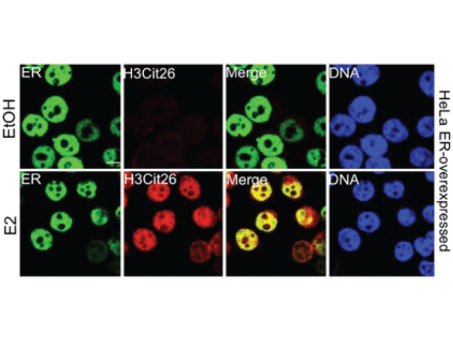
Pluripotency: Citrullination unravels stem cells
Maintenance of the pluripotent stem cell state is regulated by the post-translational modification of histones...
FULL MANUSCRIPT