Structure and Function of Membrane Bound Enzymes
STAFF: C.D. Stout, H. Heaslet, M. Yamaguchi, V.M.M. Luna, A. Annalora, J. Chartron, V. Sundaresan
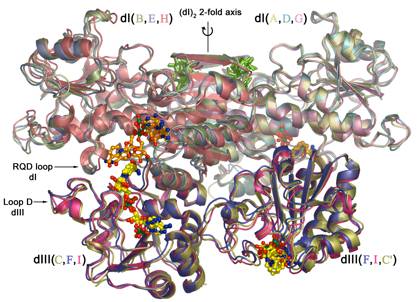
Our research is focused on the structure and function of membrane bound enzymes, and the development of methods for membrane protein crystallization. We study the mechanism of transhydrogenase, a mitochondrial respiratory enzyme complex that couples proton translocation with hydride transfer. We employ x-ray crystallography, biochemical and spectroscopic methods, electron microscopy in collaboration with M. Yeager, and NMR in collaboration with J. Dyson. Crystal structures of TH soluble domains, alone and in complex, have been determined (Fig. 1). At present the primary effort is to determine the structure of the intact, 200 kD enzyme in its membrane bound configuration.
In parallel with these studies, we are developing the application of nanodiscs for biophysical studies of integral membrane proteins in collaboration with P. Dawson, and S. G. Sligar at the University of Illinois. Nanodiscs are comprised of phospholipid binding peptides that self-assemble into discrete, water soluble, bilayer containing particles. Integral membrane proteins incorporated into these particles retain their enzymatic activity, are amenable to biochemical assays, and may have superior properties for crystallization in the absence of detergents. Both transhydrogenase and cytochrome ba3 oxidase have been incorporated into nanodiscs.
In collaboration with J. A. Fee, we are embarking on a mechanistic study of cytochrome ba3 oxidase, the terminal enzyme of respiration. The high resolution structure of the enzyme from Thermus thermophilus, crystallized in the presence of a detergent, has been determined. Crystallographic experiments, in concert with mutagenesis and spectroscopy, can visualize intermediates in the reduction of O2 to H2O, and define the paths of O2 and protons into the active site.
In collaboration with E. F. Johnson, J. R. Halpert at the University of Texas, and others, we are characterizing structures of mammalian cytochrome P450s. These membrane associated enzymes are involved in the biosynthesis lipophilic hormones, and specifically metabolize a remarkable diversity of exogenous compounds and drugs. Over 60 P450 genes occur in the human genome. High resolution structures, including substrate and inhibitor complexes, have been determined for P450s 1A2, 2C5, 2C8, 2C9, 2A6, 2A13, 3A4, and 2B4. For the latter P450 three structures of the enzyme in markedly different conformations provide insight to substrate binding and membrane insertion.
A major effort to determine the basis of HIV resistance to antiviral drugs is on-going with A. Olson, B. E. Torbett, and J.E. Elder at Scripps, and D.E. McRee at ActiveSight. One aspect of this project entails crystal structure determination of HIV protease resistant mutants with a wide range of inhibitors. Additional research projects involve crystallographic collaborations on iron-sulfur enzymes (APS reductase, with K.S. Carroll at U.C. Berkeley), electron transfer proteins (with J. A. Fee), and synthetic, self-assembling peptides (with R. Ghardiri).
Fig. 1. Superposition of three heterotrimers of transhydrogenase soluble domains observed in cocrystals. The presence of additional copies of the smaller soluble domain (dIII, lower right) in the crystal lattice provides a possible model for the intact enzyme in the membrane. PUBLICATIONS
Yadav, M.K., Redman, J.E., Leman, L.J., Alvarez-Gutierrez, J.M., Zhang, Y., Stout, C.D., Ghadiri, M.R. Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides. Biochem. 44:9723, 2005.
Carroll, K.S., Gao, H., Chen, H., Stout, C.D., Leary, J.A., Bertozzi, C.R. A Conserved Mechanism for Sulfonucleotide Reduction. Pub.Lib. of Sci. Biol 3:e250, 2005.
Yano, J.K., Hsu, M.-H., Griffin, K.J., Stout, C.D., Johnson, E.F. The structure of human microsomal cytochrome P450 2A6 with coumarin and methoxsalen bound. Nature Struct. Mol. Biol. 12:822, 2005.
Johnson, E.F., Stout, C.D. Structural Diversity of Human Xenobiotic-Metabolizing Cytochrome P450 Monooxygenases. Biochem. Biophys. Res. Comm. 338:331, 2005.
Hillier, B.J., Sundaresan, V., Stout, C.D., Vacquier, V.D. Expression, purification, crystallization and preliminary X-ray analysis of the olfactomedin domain from the sea urchin cell-adhesion protein amassin. Acta Cryst. F62:16, 2006.
Zhao, Y., White, M.A., Muralidhara, B.K., Sun, L., Halpert, J.R., Stout, C.D. Structure of Microsomal Cytochrome P450 2B4 Complexed with the Antifungal Drug Bifonazole: Insight into P450 Conformational Plasticity and Membrane Interaction. J. Biol. Chem. 281:5973, 2006.
Heaslet, H., Kutilek, V., Morris, G.M., Lin, Y-C., Elder, J.H., Torbett, B.E., Stout C.D. Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3. J. Mol. Biol. 356:967, 2006.
Yadav, M.K., Leman, L.J., Price, D.J., Brooks, C.L., Stout, C.D., Ghadiri, M.R. Coiled Coils at the Edge of Configurational Heterogeneity. Structural Analyses of Parallel and Antiparallel Homotetrameric Coiled Coils Reveal Configurational Sensitivity to a Single Solvent-Exposed Amino Acid Substitution. Biochem. 45:4463, 2006.
Chartron, J., Carroll, K.S., Shiau, C., Gao, H., Leary, J.A., Bertozzi, C.R., Stout, C.D. Substrate Recognition, Protein Dynamics, and Novel Iron-Sulfur Cluster in Pseudomonas Aeruginosa Adenosine Phosphosulfate Reductase, Nature Struct. Mol. Biol., submitted, 2006.
KEYWORDS: Transhydrogenase Cytochrome P450 Membrane proteins Crystallography