Membrane Bound Enzymes and Structure Based Drug Design
Staff: R. Akhouri, A. Annalora, Y. Chen, J.A. Fee, J. Leung, B. Liu, V.M.M. Luna, M. Yamaguchi, C.D. Stout
We use crystallography to study the structure and function of membrane bound enzymes, and to guide drug design in cytochrome P450s and HIV protease. A major focus is on transhydrogenase (TH), a mitochondrial enzyme and large integral membrane protein that utilizes the proton motive force to generate NADPH; TH mutations are correlated with type 2 diabetes. An effort to crystallize TH has progressed significantly with engineering of the maltose binding protein fusion of E. coli TH: this construct expresses extremely well, enforces correct assembly of the TH complex, affords one-step purification with efficient detergent exchange, is monodisperse, and redistributes soluble protein mass for crystallization. The TH project entails collaboration with Q. Zhang in employing novel detergents, and J. A. Tainer for solution characterization by small angle X-ray scattering.
In collaboration with J. A. Fee and staff at the Stanford Synchrotron Radiation Lightsource (SSRL), we are engaged in mechanistic study of cytochrome ba3 oxidase, the terminal enzyme of respiration. Crystal structure analysis, site-directed mutagenesis, and spectroscopic methods in solution and crystals are employed to visualize intermediates in the reduction of O2 to H2O, and to define the pathways for protons, electrons, and O2 into the membrane intercalated active site reaction chamber.
In collaboration with E. F. Johnson, J. R. Halpert at UCSD, and I. Pikuleva at Case Western, we are working toward structure determination of all key mammalian microsomal cytochrome P450s. These membrane enzymes function as a 'chemical immune system' to metabolize a vast array of exogenous compounds and drugs; knowledge of their structures is of great interest to the pharmaceutical industry. High resolution structures determined include human liver and lung P450s 1A2, 2A6, 2A13, 2C8, 2C9, 2C19, 3A4, brain specific CYP46A1, and rabbit liver 2C5 and 2B4; for the latter, five distinct inhibitor complexes display conformations from wide-open and membrane inserted to fully closed. Structures in progress include P450s 1B1, 2B6, and 2D6, and the key steriodogenic mitochondrial enzyme, CYP11A1.
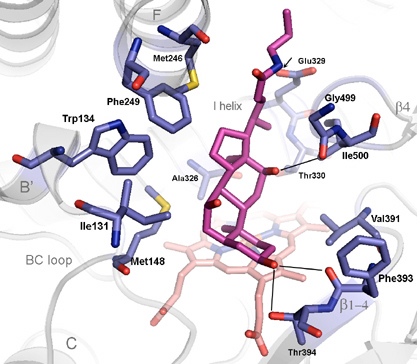
Together with A. Annalora the first mitochondrial P450 structure, CYP24A1, has been solved at 2.0 Å resolution. CYP24A1 plays a critical endocrine function by degrading the active form of vitamin D3, modulating the hormone's function in calcium homeostasis, cellular growth and apoptosis. The structure captures an open form, defining the pathway for lipophilic substrates into the active site, and reveals signature motifs for membrane insertion and interaction with the redox partner, Adrenodoxin. Active site details (Fig. 1) map out key residues that define the exquisite vitamin D specificity of this enzyme. Experiments in progress are directed toward solving closed forms with vitamin D analogs bound, and obtaining structures of CYP24A1 complexes with specific inhibitors developed at Cytochroma, Inc., now in clinical trials for chronic kidney disease.
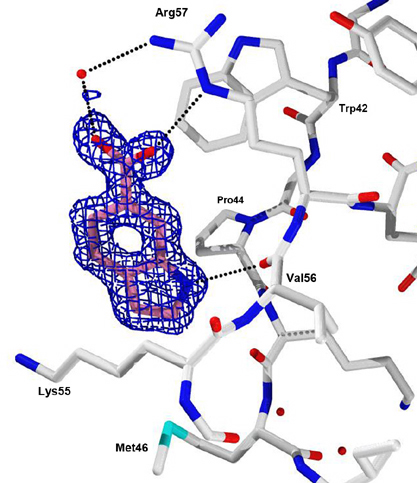
A Program Project at Scripps directed by A. Olson seeks to understand mechanisms of HIV resistance by focusing on the viral protease. With J. E. Elder our current experiments entail fragment based screening by soaking or co-crystallizing HIV protease with fragments in several crystal forms. (Fragments are small molecules about 1/3 the size of typical drugs.) The goal is to develop new classes of inhibitors that target allosteric sites outside the protease active site. Using the Active Sight fragment library and high-throughput structural genomics technologies at SSRL, we have evaluated ~1000 crystals and obtained ~400 high resolution structures. These data have yielded four 'hits', i.e. fragments binding in either of two distinct allosteric sites on HIV protease, where the active site is already occupied by a known inhibitor (Fig. 2). The next stage of the experiments will optimize these 'hits' into larger, tighter binding 'lead' molecules.
Fig. 1. Thirteen residues in the active site of the open form of the human mitochondrial cytochrome P450, CYP24A1, involved in recognition of CHAPS, a detergent of crystallization. CHAPS (magenta) is chemically similar to the natural substrate, 1,25-dihydroxyvitamin D3. Amino acid side chains are gray, and the enzyme's heme group is pink.
Fig. 2. Electron density at 1.2 Å resolution for a fragment, indole-6-carboxylic acid, bound to an allosteric site in HIV protease. Binding of this or other 'hit' fragments induces unique crystal forms of the protease when the enzyme is already in complex with the active site inhibitor, TL-3. Publications
Schoch, G.A., Yano, J.K., Sansen, S., Dansette, P.M, Stout, C.D., Johnson, E.F. Determinants of Cytochrome P450 2C8 Substrate Binding: Structures of Complexes with Montelukast, Troglitazone, Felodipine, and 9-cis-Retinoic Acid. J. Biol. Chem. 283:17227, 2008.
Giffin, M.J., Heaslet, H.A., Brik, A., Lin, Y.-C., Cauvi, G., Wong, C.-H., McRee, D.E., Elder, J.H., Stout, C.D., Torbett, B.E. AB2, an azide-alkyne click compound, is a potent protease inhibitor of multidrug-resistant HIV-1. J. Med. Chem. 51:6263, 2008.
Mast, N., White, M.A., Bjorkhem, I., Johnson, E.F., Stout, C.D., Pikuleva, I.A. Crystal structures of substrate bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain. Proc. Natl. Acad. Sci. USA 105:9546, 2008.
Gay, S.C., Sun, L., Maekawa, K., Halpert, J.R., Stout, C.D. Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand induced structural response through α-helical repositioning. Biochemistry:in press, 2009.
Pornillos, O., Ganser-Pornillos, B.K., Kelly, B.N., Hua, Y., Whitby, F.G., Stout, C.D., Sundquist, W.I., Hill, C.P., Yeager, M. X-ray structures of the hexameric building block of the HIV capsid, Cell:in press, 2009.
Keywords
Transhydrogenase
Cytochrome oxidase
Cytochrome P450
Membrane proteins
HIV protease
Crystallography
Fragment screening