 Figure 1. Improved shape potentials
Figure 1. Improved shape potentials
 Figure 2. Basis for improved electrostatic potential
Figure 2. Basis for improved electrostatic potential
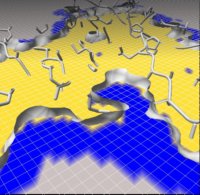 Figure 3. The crystallographic structure of the nucleosome core particle (NCP).
Figure 3. The crystallographic structure of the nucleosome core particle (NCP).
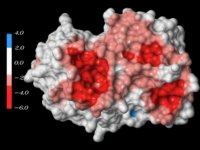 Figure 4. The electrostatic potential of the histone core of the NCP
Figure 4. The electrostatic potential of the histone core of the NCP
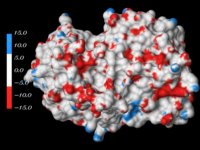 Figure 5. DOT global search results for B-DNA fragments docked to the histone core
Figure 5. DOT global search results for B-DNA fragments docked to the histone core