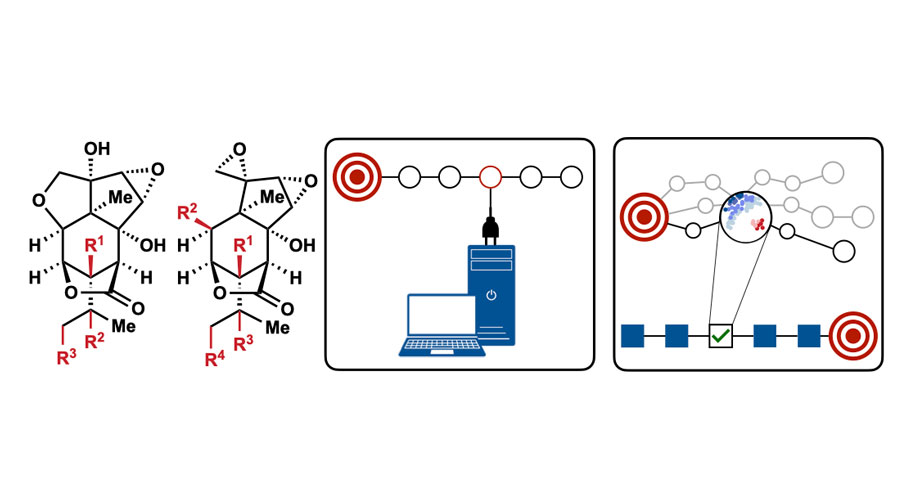
Scripps Research scientists used computational tools to determine how to synthesize picrotoxanes (left). The tools helped analyze the behavior of intermediate molecules (right). Credit: Scripps Research.
Virtual chemistry speeds up drug discovery
By using computer modeling to predict chemical reactions, Scripps Research scientists were able to synthesize 25 variations of a plant compound that could treat brain diseases.
January 03, 2025
LA JOLLA, CA—Among the hundreds of thousands of chemical compounds produced by plants, some may hold the key to treating human ailments and diseases. But recreating these complex, naturally occurring molecules in the lab often requires a time-consuming and tedious trial-and-error process.
Now, chemists from Scripps Research have shown how new computational tools can help them create complex natural compounds in a faster and more streamlined way. They used their approach, described in Nature on December 23, 2024, to synthesize 25 different picrotoxanes, compounds which are originally found in plants and have the potential to alter pathways in the brain.
“It has been incredibly hard to manipulate these types of complex plant compounds for the benefit of drug design,” says senior author Ryan Shenvi, PhD, professor at Scripps Research. “The ability to now combine virtual predictions with real-world experimentation marks a turning point in how we design and build molecules.”
Picrotoxanes, found in the seeds of certain Asian and Indian shrubs, are known to affect the mammalian nervous system; they bind to the same brain receptors targeted by the anxiety and sleep medication Valium. They have been used in some cultures as pesticides or to kill fish. Because these compounds can be ingested orally and affect brain function, researchers like Shenvi have been interested in whether they might have therapeutic potential. However, scientists have only been able to make a few picrotoxanes in the lab, making it hard to manipulate and study them.
“Like many other plant metabolites, the atoms of picrotoxanes are arranged in a complex way that makes their behavior hard to predict,” says Shenvi. “We could not assume that a reaction that worked to synthesize one picrotoxane would work on another, even if it looked almost identical.”
Shenvi and Chunyu Li, a Scripps Research graduate student, were struggling to synthesize picrotoxanes and turned to advanced computer modeling to predict new ways to create picrotoxanes from basic chemical building blocks. They first generated a virtual library of possible intermediate compounds that could be formed during the synthesis of picrotoxanes. Then, they used a model known as Density Functional Theory (DFT) to analyze the behavior of these intermediates, flagging those that were likely to both succeed and quickly lead to neuroactive compounds.
When the group tested five picrotoxane synthesis pathways suggested by the modeling — three that were predicted to succeed and two predicted to fail — all five outcomes were correct.
“DFT is commonly used post-hoc, to explain experimental data and how a chemical reaction works, so I was skeptical that it would work in this predictive way,” says Shenvi. “And I was shocked when it worked so well.”
DFT, however, is still relatively time-consuming to use for each possible intermediate. Shenvi and Li wanted to scale their approach up and make it even faster to create more picrotoxanes. They used a pattern-recognition technology similar to the one underlying many modern artificial intelligence (AI) programs in order to find patterns in the DFT results. They were able to create a new statistical model that predicted reaction success in a fraction of the time. Using that model, they identified synthesis techniques for 25 picrotoxanes and showed in the lab that they worked.
“This approach didn’t just let us create picrotoxanes,” says Li. “It paves a way for chemists to solve other difficult synthesis problems.”
Shenvi says the lab is already applying the approach to other problems. They are also planning to continue testing the 25 picrotoxanes they can now produce to see how they impact mammalian biology.
Shenvi and Li are the authors of the study, “Total synthesis of twenty-five picrotoxanes by virtual library selection.”
This work was supported by funding from the National Institutes of Health (GM122606) and a Skaggs Graduate School of Chemical and Biological Sciences Dale Boger Endowed Graduate Fellowship.
For more information, contact press@scripps.edu

