New electron microscopy technique could shortcut the development of vaccines and monoclonal antibody therapies
The new method by scientists at Scripps Research identifies specific antibodies in immune responses to vaccination or infection in fraction of time needed for traditional approach.
January 19, 2022
LA JOLLA, CA—Scientists at Scripps Research have devised a method that may be able to shortcut one of the big steps in modern vaccine development.
The researchers, whose work appears in Science Advances on January 19, 2022, showed that they could use high-resolution, low-temperature electron microscopy (cryo-EM) to rapidly characterize antibodies—elicited by a vaccine or infection—that bind to a desired target on a virus at an atomic level.
“The COVID-19 pandemic has highlighted the need for robust and rapid vaccine and antiviral technologies,” says study senior author Andrew Ward, PhD, a professor in the Department of Integrative Structural and Computational Biology at Scripps Research. “We are optimistic that our new approach will help fill that need by greatly streamlining antibody discovery.”
“Traditionally, identifying antibodies that are useful against a virus involves the laborious sorting and testing of antibody-producing B cells to find the right ones—a process that takes months,” says Postdoctoral Research Associate and study co-first author Aleksandar Antanasijevic, PhD.
“With this new method we can go from blood sample collection from infected or immunized patients to identifying all the elicited antibodies of interest in about ten days,” adds Staff Scientist and study co-first author Charles Bowman.
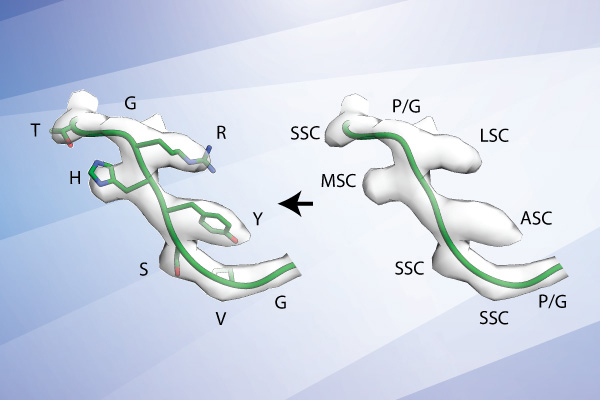
A new method developed at Scripps Research uses cryoEM technology to more quickly identify antibodies for use in vaccine development. In this example, the algorithm screened between approximately 100,000 to 1,000,000 possible antibody sequences from the database to identify the sequence (left) that best matches the antibody observed in their cryoEM images (transparent gray surface).
The researchers’ feat was enabled in part by recent improvements in cryo-EM, a technology that uses a beam of electrons to illuminate and image targets far below the scale of ordinary light microscopy. In a study published in Nature Communications in August, for example, the researchers used high-resolution cryo-EM to rapidly and precisely map where antibodies in rhesus macaque monkeys bind to synthetic versions of the HIV envelope protein that are being developed for potential HIV vaccines.
In the new study, the team took this line of research a step further. They employed a “structure-to-sequence” computer algorithm that can relate antibody structure determined by cryo-EM, to the DNA sequence that would produce that structure. For this, they assembled a library of all the antibody-encoding DNA sequences from the monkeys—obtained by quickly bulk sequencing the genetic material from antibody-producing B-cells from the animals’ lymph nodes. Applying the algorithm to the cryo-EM data and the antibody sequence library, the scientists could reliably match an antibody of interest in their cryo-EM images to a unique antibody defined in the sequence database.
The researchers showed that they could confirm the accuracy of the result by making copies of that unique, “monoclonal” antibody using the sequence data, and verifying with cryo-EM that the antibody binds in an identical way to the originally imaged antibody.
The scientists are now refining their technique to optimize its speed and usability, and are applying it to several areas: to rapidly evaluate human antibody responses to experimental HIV vaccines; to develop antibody-blocking treatments for autoimmune diseases; and to discover antibodies that could therapeutically hit other protein targets on cells.
They expect that future improvements in cryo-EM technology and structure-to-sequence algorithms will allow the even more rapid identification of antibodies using high-resolution structural images alone, with no need for DNA sequencing of B cells.
“This structure-to-sequence approach has a lot of potential in immunology and beyond,” Antanasijevic says. “We envision being able to use it someday to study protein-to-protein interactions generally, for example, to discover a given protein’s binding partners.”
“ From Structure to Sequence: Antibody discovery using cryoEM” was co-authored by Aleksandar Antanasijevic, Charles Bowman, Christopher Cottrell, Gabriel Ozorowski, Leigh Sewall, Bartek Nogal, Fangzhu Zhao, Bettina Groschel, William Schief, Devin Sok, and Andrew Ward, all of Scripps Research; by Robert Kirchdoerfer of the University of Wisconsin—Madison; by Amit A. Upadhyay, Diane Carnathan, Chiamaka Enemuo, Guido Silvestri and Steven Bosinger of Emory University; and by Kimberly Cirelli and Shane Crotty of La Jolla Institute for Immunology.
The research was supported by the National Institutes of Health (UM1AI100663, UM1AI144462, P01 AI110657), the Bill and Melinda Gates Foundation through the Collaboration for AIDS Vaccine Discovery, and the amfAR Mathilde Krim Fellowship in Biomedical Research, among other sources.
For more information, contact press@scripps.edu

