
Conformational Dynamics of DNA Polymerases
DNA polymerases are remarkable for their ability to synthesize DNA at rates approaching several hundred base pairs per second while maintaining extremely low error frequencies. To achieve such high replication fidelity, polymerases must discriminate between correct and incorrect substrates during each cycle of nucleotide incorporation, in accord with the ever-changing identity of the template base. In addition, the ability of the polymerase to remove misincorporated nucleotides using a separate 3’-5’ exonuclease activity (proofreading) further enhances the overall fidelity of the enzyme. These fundamental aspects of polymerase function are intrinsically dynamic and require large conformational changes and rearrangements of the enzyme-DNA-nucleotide complex. The rate-limiting step in nucleotide incorporation involves a transition from an “inactive” to “active” ternary complex, which is thought to involve movement of the fingers subdomain of the polymerase. However, such motions have yet to be observed directly in solution. During proofreading, the 3’ terminus of the newly synthesized DNA must travel some 35 angstroms from the polymerase (pol) active site to a separate 3’-5’ exonuclease (exo) site. The mechanisms by which this movement is achieved and regulated are currently unknown. We are using single-molecule fluorescence techniques to monitor the dynamics of nucleotide incorporation and to observe switching of the DNA substrate between pol and exo sites of the Klenow fragment of E. coli Pol I. Large-scale movements of protein domains or of the DNA substrate are observed by means of Förster resonance energy transfer (FRET) between strategically placed donor and acceptor groups. These studies are providing insights into the dynamic conformational transitions that underlie DNA polymerase function.
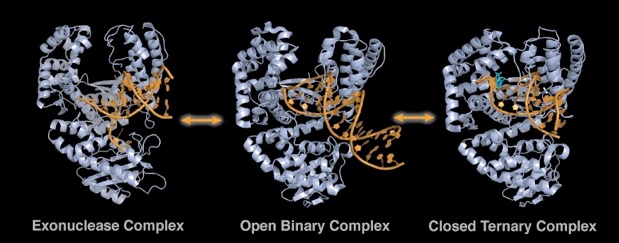
Figure 1. Replicating and editing complexes of DNA polymerase I. The center structure is the inactive (open) complex of Bacillus Klenow fragment (grey) bound to a primer-template (blue, PDB ID: 1L3V). Following addition of the correct nucleotide (orange) the finger domain closes to generate an active replicating complex (right structure, PDB ID: 1LV5). If the primer strand posses a 3’ mismatch, the primer relocates from the polymerizing active site to the exonuclease site (left structure, PDB ID: 1KLN (E. coli)).

Figure 2. Single-molecule analysis of Klenow fragment interactions with a primer-template utilizing total internal reflection fluorescence (TIRF). A) Time traces of anti-correlated donor (green) and acceptor (red) signals (top) and corresponding FRET efficiency trace (yellow) and fit line (white) (bottom). B) FRET efficiency histogram. C) 2-D contour plot of starting verse stopping FRET efficiencies. D) Dwell time histogram and corresponding dissociation rate.
Recent Millar Laboratory Publications
S.Y. Berzhna, J.P. Gill, R. Lamichhane and D. P. Millar, "Single-Molecule Förster Resonance Energy Transfer Reveals an Innate Fidelity Checkpoint in DNA Polymerase I ", Journal of the American Chemical Society 134, 11261-1268 (2012).
J. Gill, J. Wang and D. Millar, "DNA Polymerase Activity at the Single-Molecule Level", Biochem. Soc. Trans. 39, 595-599 (2011).
Stengel, G., Gill, J. P., Sandin, M., Wilhelmsson, B., Albinsson, B.N. and Millar, D.P. (2007) Conformational Dyanmics of DNA Polymerase Probed with a Novel Fluorescent DNA Base Analogue. Biochemistry, 46, 12289-12297.
PDB References
1KLN. Beese, L.S., Derbyshire, V. and Steitz, T.A. (1993) Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science, 260, 352-355.
1L3V and 1LV5. Johnson, S.J., Taylor, J.S. and Beese, L.S. (2003) Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A., 100, 3895-3900.
