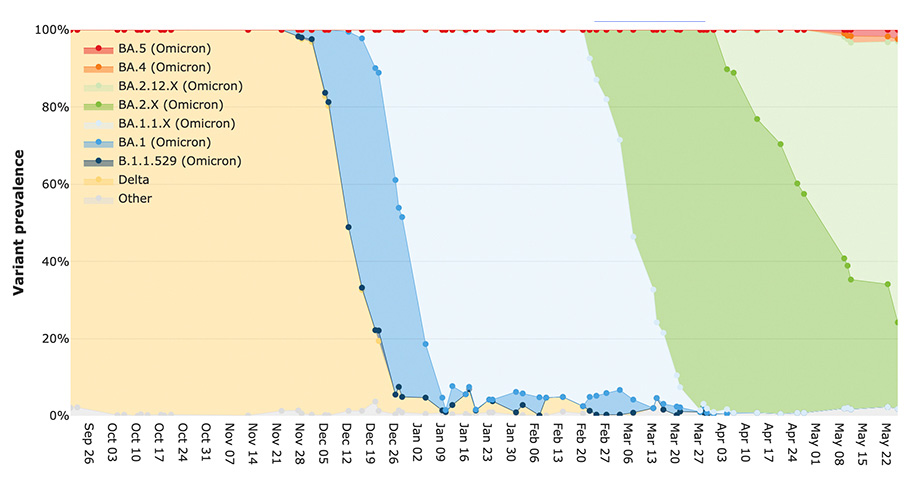
Researchers devised a new way of analyzing wastewater to monitor the prevalence of different SARS-CoV-2 variants over time (shown here for San Diego, from September 2021 to May 2022). Credit: SEARCH alliance
Scientists develop tools for early detection of SARS-CoV-2 variants in wastewater
New tools developed at Scripps Research and UC San Diego are helping public health officials around the world get vital information about pathogen variants from wastewater.
July 07, 2022
LA JOLLA, CA—It can be a bit smellier than other ways of monitoring COVID-19, but analyzing wastewater is a cheaper, faster and more accurate way for public health officials and researchers to detect rising cases. Bits and pieces of the SARS-CoV-2 virus are flushed down toilets and washed down sinks by infected individuals; more copies of the virus found in sewage means more people are sick. But until now, most wastewater analysis methods lumped all SARS-CoV-2 viruses together as one.
Now, scientists at Scripps Research and University of California, San Diego, in collaboration with the San Diego Epidemiology and Research for COVID Health (SEARCH) alliance, have changed that. The team has reported that with just two teaspoons of raw sewage, they can accurately determine the genetic mixture of SARS-CoV-2 variants present within a population and identify new variants of concern up to 14 days before traditional clinical testing. In San Diego wastewater, the group detected the Omicron variant 11 days before it was first reported clinically.
Their algorithm, named “Freyja”, for identifying SARS-CoV-2 variants in wastewater, described today in Nature, has quickly been adapted by many public health labs, and is a boon to surveillance efforts that aim to detect new variants of SARS-CoV-2.
“In a lot of places, standard clinical surveillance for new variants of concern is not only slow but extremely cost-prohibitive,” says Kristian Andersen, PhD, professor of Immunology and Microbiology at Scripps Research and a senior author of the new work. “But with this new tool, you can take one wastewater sample and basically profile the whole city.”
The project required a tight collaboration between hospitals, state and local governments, sequencing facilities, and academic scientists–including researchers in the Andersen lab and that of UC San Diego microbiologist Rob Knight, PhD. The Knight lab deployed 131 wastewater autosamplers to collect wastewater from 343 buildings on the UCSD campus and 17 public schools across 4 San Diego school districts, and acquired samples from large wastewater treatment facilities in the county. Over the course of nearly a year, the group analyzed more than 20,000 wastewater samples. In the process, they developed improved methods for concentrating viral RNA in wastewater, which are now being widely used by public health labs across the country and world. Then, the Andersen lab took on the challenge of quantifying viral variants from the sequencing data.
“It’s challenging to take all these tiny pieces of virus floating around in wastewater and figure out which ones are from different variants and what their relative abundance is,” says Scripps Research postdoctoral fellow Joshua Levy, PhD, a co-first author of the new paper with Smruthi Karthikeyan of UC San Diego.
Many variants of SARS-CoV-2, including Omicron and Delta, differ by a small number of mutations. But since these changes can impact how the virus spreads or infects people, public health officials must carefully track them. They have typically done this by sequencing virus genomes from patients, which is a slow and costly process and has become less effective at capturing the extent and diversity of COVID-19 variants as many people turn to at-home testing.
Levy developed a library of “barcodes” that identify SARS-CoV-2 variants based on short snippets of their RNA that are unique to each variant. Then, he coded a new computational tool that sifts through the mass of genetic information in wastewater to find these barcodes. He made the new Freyja program easy to use and free.
“If you’re in a lab that can already sequence a wastewater sample, you’re good to go— you just run this code and in another 20 seconds you’re done,” he says.
When the researchers applied Freyja to their wastewater samples and compared the results to clinical data collected from around San Diego by SEARCH, they discovered that the tool detected variants of concern, including Alpha, Delta, and Omicron, in wastewater up to 14 days before it was reported clinically. The Mu (B.1.621) variant was detected in UC San Diego wastewater on July 27, 2021— four weeks before its first clinical detection on campus. And, using more recent data not included in the original study period, the team also reported that the Omicron variant could be detected at the Point Loma wastewater treatment plant—at an abundance of just over one percent of all SARS-CoV-2 viruses in a contributing population of over two million people— on November 27, 2021, 11 days before its clinical detection in the city.
“Wastewater contains a massive amount of very valuable information about our health, including these viral genomes that can let us track the course of a pandemic or epidemic,” says Karthikeyan.
“It took a lot of collaboration between public health and academic players to get this system established in San Diego, and now that we’ve shown its effectiveness, we hope it inspires other localities to use these tools,” adds Knight. “We’re also very excited about expanding them to pathogens beyond SARS-CoV-2.”
The researchers say they are continuing to improve upon the set of tools they use to analyze viruses in wastewater, but that the current suite of methods is already a leap forward from previous approaches. The same strategies could be used to not only track variants of SARS-CoV-2 but other human pathogens.
“When you rely on clinical sampling, you not only introduce a lot of socioeconomic and geographic bias into who is contributing to genomic surveillance data, but you also have the problem of asymptomatic people not getting tested and those who only use home tests not contributing to the pool of data,” says Levy. “But with wastewater, we don’t have those blind spots.”
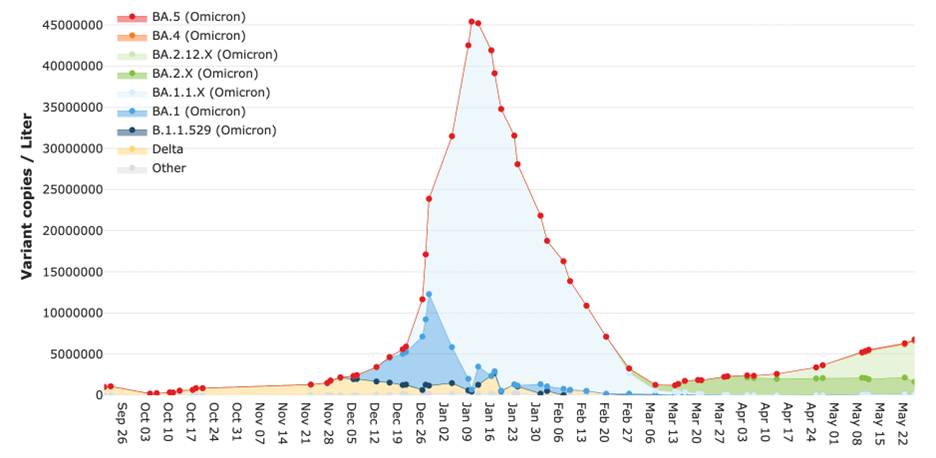
Using wastewater virus concentration measurements and genome sequencing, researchers tracked the rapid spread of SARS-CoV-2 during the Omicron wave of the pandemic. Credit: SEARCH alliance
In addition to Levy, Karthikeyan, Andersen and Knight, authors of the study, “Wastewater sequencing uncovers early, cryptic SARS-CoV-2 variant transmission,” include Christine Aceves, Catelyn Anderson, Karthik Gangavarapu, Emory Hufbauer, Ezra Kurzban, Justin Lee, Nathaniel Matteson, Edyth Parker, Sarah Perkins, Karthik Ramesh, Refugio Robles-Sikisaka, Madison Schwab, Emily Spencer, Shirlee Wohl, Laura Nicholson, and Mark Zeller of Scripps Research, as well as collaborators at UC San Diego, Rady Children’s Institute for Genomic Medicine, Scripps Health, Sharp Healthcare, Helix, the County of San Diego Health and Human Services Agency, the California Department of Public Health, and the Centers for Disease Control and Prevention.
This work was supported by funding from the U.S. Centers for Disease Control and Prevention (75D30121P10258, 75D30120C09795), National Institutes of Health (5T32AI007244-38, 1DP1AT010885, S10OD026929), National Institute of Allergy and Infectious Diseases (3U19AI135995-03S2, U19AI135995, U01AI151812), National Center for Advancing Translational Sciences (UL1TR002550), the Conrad Prebys Foundation, National Science Foundation (RAPID 2029069), and the San Diego County Health and Human Services Agency.
For more information, contact press@scripps.edu

